Codon bias
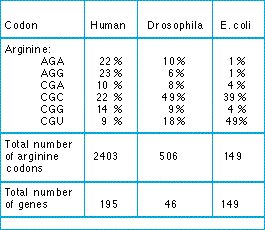
Biases in codon usage provide evidence for constraints on silent sites. If all the silent alternative codons were all functionally equivalent, we should expect only random variation in the frequency of those codons in a species. The table shows that there are, in fact, consistent biases.
How selection could discriminate among silent codons:
• The nucleotide sequence controls the secondary structure of the DNA molecule; changes in nucleotides might then influence the molecular shape, which could make a difference to the organism's fitness. Silent substitutions would be as likely to influence structure as replacement ones, and selection would work on both for the same reason.
• The different silent codons use different tRNA molecules. It has been observed, in yeast and in Escherichia coli , that the frequency of use of the codons in a set of synonyms is correlated with the tRNA abundances in the cell. We should expect the relation if the frequencies of codons were set by some other factor: the tRNA abundances would then adjust (by natural selection) to the quantity needed. It has also been suggested that codon biases are caused by directional (or biased) mutation.
Table: frequencies of six arginine codons in the DNA of three species. The table gives the percentages of arginine amino acids that are encoded by each of the six codons in various numbers of genes in the species. From Grantham et al. (1986).
| Next |