The reconstruction of phylogeny - What are the different molecular methods?
The problem with distance statistics
The problem with distance statistics is that they do not distinguish the three different kinds of shared character. If shared ancestral similarities are more common between two species than are shared derived similarities, distance statistics confuse paraphyletic and monophyletic groups. If some members of a group of species (like the reptiles) evolve slowly while other members of the group evolve rapidly, the slowly evolving species are left looking similar even though they do not share a recent common ancestor; distance statistics group species by similarity and will recognize the disparate set of slowly evolving species as a phylogenetic group.
Does this general problem apply to DNA hybridization?
DNA hybridization measures the percentage similarity of the whole (or a large part of) the DNA of two species. The DNA sequences of species may well evolve at a relatively constant rate. Genes and proteins provide reasonably good molecular clocks and the same is likely to be true for the DNA as a whole. Then the DNA hybridization method is justified. The problematic cases like reptiles arise when evolution proceeds more rapidly in one lineage than another. But if that rapid evolution occurs only in some characters, such as observable morphology; and if other characters, such as the changes in the whole DNA molecule measured by hybridization, evolve at a constant rate, distance statistics can be reliable for the constantly evolving characters.
In summary, there are classes of molecular evidence, of which DNA - DNA hybridization is currently the most important, that can be used to infer phylogenies by distance statistics. Distance statistics suffer the general disadvantage that they do not concentrate on shared derived homologies. They are most reliable if molecular evolution is clock-like, which in turn is most likely if it driven by neutral drift.
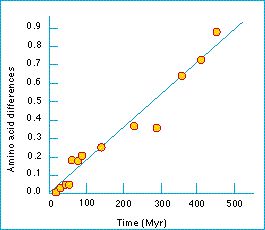
Figure: the rate of evolution of hemoglobin is constant enough to be described as a molecular clock. Each point on the graph is for a pair of species, or groups of species. From Kimura (1983).
| Next |